James Cameron’s descent to the Challenger Deep – we have adventure, intrigue, and a great story for the media. But we also have an amazing opportunity for SCIENCE!
Despite a faulty hydraulics hampering sample collections, the Deepsea Challenger managed to grab half a sediment core – a cupful of muddy, watery ooze from the deepest point in the ocean:
“Jim recovered about 50 millileters of muddy seawater that I gleefully processed for culturing and for genomic studies,” Doug Bartlett, chief scientist for the DEEPSEA CHALLENGE project, said in an email to National Geographic News.
“Can’t wait to see what new critters (Bacteria, Archaea, and fungi) that we discover,” said Bartlett, a marine biologist at the Scripps Institution of Oceanography in San Diego, California.
Some might lament Cameron’s technical difficulties and shake their heads at the lost sampling opportunity. But even half a sediment core will reveal precious information about one of the last frontiers on earth. We have plenty to work with.
In molecular terms, 50 milliliters is a LOT of sample. Normally my lab protocols call for 200 microliters of mud for a single extraction of environmental DNA. So with his one cup of mud, James Cameron can do 250 DNA extractions–and you only need one or two extractions (maybe a few more, which are then concentrated and pooled if there isn’t a lot of DNA because of few animals or small amounts of tissue) before you can move forward and produce gene sequences, using high-throughput platforms such as the Illumina Hi-Seq.
So even with a single drop of sample, you can obtain hundreds of millions of DNA sequences from species inhabiting the Challenger Deep. And there’s no restriction to any particular taxonomic group. The power of DNA means that we will be able to characterize deep sea life across all known domains–bacteria, archaea, eukaryotes, and even viruses.
One of the first things to sequence will be ribosomal RNA, a conserved gene that essentially serves as a molecular barcode (since every cell needs its ribosomes to survive!) and allows us to place species on branches within the Tree of Life. By comparing ribosomal genes from the Challenger Deep to those from species that have already been studied, we’ll be able to place this deep-sea community in an evolutionary context and investigate how life might have evolved in the ocean depths. What other environments contain closely related species? How divergent are the ribosomal genes in the Mariana Trench (and from this, we can start guessing how long these trench communities have been isolated–if at all–from other deep sea habitats)? Is the Challenger Deep harboring any novel, undiscovered branches on the Tree of Life?
We’ll also get an environmental metagenome from this sequencing effort — randomly sequenced pieces of DNA representing every species’ genome lurking in that muddy sample. This will give us an expanded view compared to ribosomal genes, and we can start inferring things about community function. What type of genes are prevalent in the deepest, darkest ocean trench? The types of genes we find can tell us a lot about how a community survives (does it rely on scarce food sinking from above, or have species adapted to use alternative metabolic pathways such as chemosynthesis), and how an assemblage of organisms might inherently depend on each other to survive in an extreme environment. If the community in the Challenger Deep is not too complex (a handful of species, or a good pool of abundant ones) and the scientists at Scripps decide to sequence a LOT of DNA from this precious mud (a couple runs on the Illumina Hi-Seq can get you close to a billion DNA sequences), then it is possible that we might be able to assemble whole genomes from this random sample of mud. So instead of a ribosomal gene we’ll potentially have an entire genome as a molecular barcode for some microbial species–and for inferring how evolution happened in the deep-sea, a genome will give you a lot more information than just a short ribosomal sequence.
In addition to extracting DNA we can also take out the RNA and look at patterns in molecules such as mRNA (expressed transcripts of genes, if you remember back to high school biology). So in addition to finding out who’s there and what their genomes say they can do, RNA can tell us what these species might actually be doing. Remember that there’s a lot of “junk” DNA sitting around in any given genome (and you’ll get a lot of this information from a random environmental metagenome sequencing), so its always good to have additional information about what type of genes are being expressed. Now the pressure and temperature changes will have sent most species’ cellular machinery into overdrive during Cameron’s ascent to the surface, and we may get more of a “help, help , I’m dying” reaction from the community. Its always tricky to interpret gene expression. But in many ways, any data is good data. Gene expression from the Challenger Deep may tell us some very exciting things.
Doug Bartlett from Scripps also indicated plans to try and culture some microorganisms from Cameron’s sample — while there’s no guarantee of success (the pressure change and inherent difficulty in culturing microbes both present significant hurdles), any cultured species would be closely scrutinized and provide mountains of data for years to come. Not only could we sequence genomes from cultured species, but we could organize sophisticated experiments to figure out exactly what nutrients they need, their metabolic pathways, and novel compounds produced that all contribute to adaptation in the deep sea. We may soon have alien life growing in a Southern California lab!
These approaches alone will give you an unprecedented view into life in the Mariana Trench. But we can still do more with that half core.
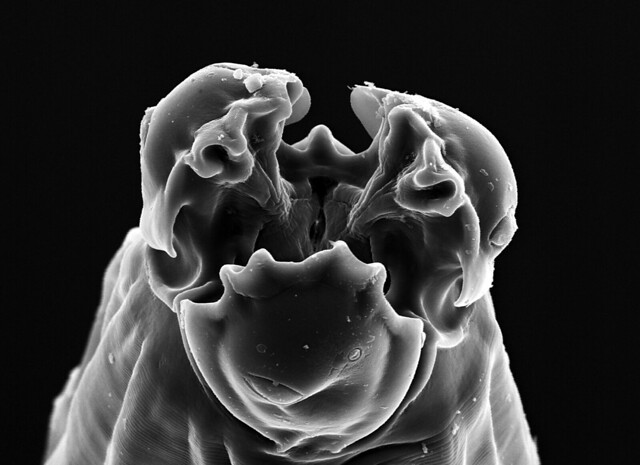
Cameron noted that the ocean floor he saw was lunar-like, smooth and featureless–but that doesn’t mean the environment is exclusively the realm of microbes. In fact, we know that bigger (albeit still microscopic) species like foraminifera do live in the Challenger Deep (Todo et al., Science, 2005), and Cameron saw amphipods swimming around before the sample had even been returned to the lab. Which means we can probably get some cool visuals if we take another drop of mud and peer at its contents under the microscope. If there are amphipods and forams, there will be nematodes in Cameron’s sample. If nematodes can live ~3km deep within the fracture water of South African mines, they can certainly put up with a little bit of pressure and scraps of food in the Hadal zone. So yeah, we should be able to get some pictures like this (for inspiration and general awesomeness):
I’m not done yet. We can still get more from that core, including:
- Characterizing the chemical makeup of the sediment. This can be done via methods such as stable isotope analysis, and we can ask questions such as: Can we pinpoint the original source of the organic matter in the Challenger Deep? What type of food is available for organisms down there? What does that say about conditions and species’ habitats in the deepest ocean trenches?
- Sediment geochemistry. What type of sediment is down there? Where did it come from (what continent or ocean region) and what trajectory might it have taken as it slowly sunk to the deepest depths?
All in all, Cameron’s sample will fundamentally contribute to our knowledge about some big questions in biology, such as:
Biological adaptations to life in extreme environments
Tim Shank, a deep-sea biologist at the Woods Hole Oceanographic Institution in Massachusetts, says that the waters above Challenger Deep are extremely unproductive; there is little algal life at the surface, and, therefore, less food is cycled down to deeper waters. “If it had been a trench with a productive water column, like the Kermadec Trench near New Zealand, I think he would have seen much more biology,” says Shank. However, sediment samples are certain to contain billions of microbes.
Insight into what life might be like on other planets:
The mud could contain exotic species of microbial life that may not only advance our understanding of the deep ocean but also help in the search for extraterrestrial life. For instance, scientists think Jupiter’s moon Europa could harbor a global ocean beneath its thick shell of ice—an ocean that, like Challenger Deep, would be lightless, near freezing, and home to areas of intense pressure. (See “Could Jupiter Moon Harbor Fish-Size Life?”)
So for those of you that scoffed at the botched sampling, there’s some serious scientific amazingness that awaits us in that half core of mud.






Good post, thank you! And the expedition and dives will likely continue to afford many more sampling opportunities. Stay tuned for more info on future sampling and research.