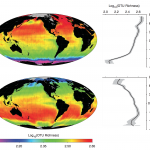
This might come as a shocker: I don’t care about metabolism (or bits of floating plastic, or whale sharks, or coral reefs…sorry Deeplings). Its not that I’m not interested – these fields are fascinating and scientifically important. But on a day-to-day basis, when I’m overloaded with data analysis, grant proposals, and a bursting inbox, I just don’t care. I can’t care. I have to focus on my immediate projects, publish papers, get a job, get tenure.
But when I read Dr. M’s new (P ***ing NAS) paper last week, it hit me: I really should care. We all should. Its not about forcing to ourselves to waste time reading about topics or environments that aren’t relevant. All knowledge is essential–especially diverse knowledge, especially in today’s changing landscape of science.
How can energy limitation in the deep-sea be relevant to a parent trying to keep his/her kitchen germ-free? It’s all relevant, because in each case we’re trying to understand how complex communities function and change over time – regardless of whether a given ecosystem resides in the built environment or a natural setting.
Biology is heading towards integrated data – I study microbial genomics, but I should also be thinking about metabolism and temperature effects on the species I search for in environmental data. For us genomicists we’re so used to dealing with so little information. We just get (rather large) computer files listing lots and lots of DNA. The challenge for us is to relate those A’s, T’s, C’s and G’s back to something meaningful. But of course we don’t just want to look at DNA – we want a holistic understanding of ecosystem function. I want to know how one piece of DNA relates to a body size, how that body size relates to the type of food that pariticular species eats, and how all that interacts on a grand scale in an ecoysytem.
Often I feel like us genomicists have our hands tied – but we have a powerful tool in our pocket (that’s what she said) that can set us free in ways that no one else can experience. DNA is objective in ways that other types of data aren’t–taxonomy is subjective and decisions vary depending on the expert identifying a specimen. What if we could use parameters like temperature, depth and type of food input to predict what species will be there? Of course, that’s a lifelong obsession for researchers like Jack Gilbert (@gilbertjacka) and colleagues, and we’re steadily making progress towards these modeling goals. Some of the new microbial model papers are pretty badass.
To summarize the ongoing transformation in biology, I’ll bestow some eloquent foresight from Poole et al. 2012:
As researchers seek to go beyond function and understand the effects of global environmental changes on ecosystems [6], metagenomics will be essential. It has already helped to unlock the mechanisms for climate–carbon-cycle feedbacks [7] and, for simple microbial ecosystems, has illuminated the probable metabolic basis for key community interactions [8]. These examples underscore two crucial points. First, genomic knowledge is increasing the understanding of how simple organisms interact with their multicellular counterparts in an ecosystem context [9]. Second, the ability to zoom in on the functional roles of species within an ecological community [3] will make metagenomics indispensable for the future study of whole-ecosystem functioning.
And this data revolution isn’t simply relegated to basic research. In terms ecotoxicology and ecosystem monitoring, Van Aggelen et al 2010 note that:
Omic and bioinformatic tools offer substantial promise for discovery of gene, protein, and/or metabolite alterations indicative of the mode of action (MOA) of chemicals and improved understanding of mechanisms in prospective studies (Ankley et al. 2006). Knowing the MOA can reduce uncertain ties in chemical risk assessments, providing, for example, a basis for extrapolating effects across species (Benson and Di Giulio 2007).
Ideally, omics data would reflect both the MOA and deleterious outcome(s). To achieve this, the cascade of pathways associated with toxicity must be defined, from a molecular initiating event (e.g., receptor binding) through subsequent biological alterations (reflected by omic and cellular changes) that culminate in a deleterious outcome (NRC 2007).
Thus, although gene expression is affected by many environmental factors, a subset of genes with altered expression can inform on stress responses.
The biology landscape (and earth’s climate) are changing and science must adapt. Scientific infrastructure and even researcher mindsets must change in order to accomodate a new order of thinking.
There is also a need to build capacity within academia, the private sector, and government agencies to implement omic tools and to evaluate omics data, particularly with respect to biological and ecological significance. These institutions will require resources, support, and targeted training to bring scientists and decision makers within their organizations to a point where these tools can be used effectively in regulatory decision making, especially in risk assessment. (Van Aggelen et al 2010)
The deep sea may face a perpetual energy recession, but in terms of scientific data we’re about to experience one hell of an economic boom.
References:
McClain CR, Allen AP, Tittensor DP, Rex MA. (2012) Energetics of life on the deep seafloor. Proc Natl Acad Sci USA. Advance Access
Poole AM, Stouffer DB, Tylianakis JM. (2012) “Ecosystomics”: ecology by sequencer. Trends in Ecology & Evolution, 27(6):309–10.
Van Aggelen G, Ankley GT, Baldwin WS, Bearden DW, Benson WH, Chipman JK, et al. (2010) Integrating omic technologies into aquatic ecological risk assessment and environmental monitoring: hurdles, achievements, and future outlook. Environ. Health Perspect. p. 1–5.